library(tidyverse)
# find data at https://doi.org/10.1002/agj2.20812
dd_yield <- readxl::read_xlsx("data/8.Yield_Plant_Measurements.xlsx", sheet = 2) %>%
filter(Year == 2014,
State == "IA",
Site == "Ames") %>%
mutate(across(VT_TissN:R4N, ~ as.numeric(.)))
dd_soil <- readxl::read_xlsx("data/1.Site_Characterization.xlsx", sheet = 2) %>%
filter(Year == 2014,
State == "IA",
Site == "Ames",
Horizon == "Ap") %>%
mutate(Block = as.numeric(Block),
across(Clay:WC, ~as.numeric(.)))
dd_N <- readxl::read_xlsx("data/4.SoilN.xlsx", sheet = 2) %>%
filter(Year == 2014,
State == "IA",
Site == "Ames",
Plot_ID %in% dd_yield$Plot_ID,
Sam_Time == "Post") %>%
mutate(across(c(Plot_ID, N_Trt, Plant_N, Side_N, Plant_N_SI, Side_N_SI), ~as.numeric(.)))
dd_complete <- dd_yield %>%
left_join(dd_soil) %>%
left_join(dd_N)
dd_complete <- dd_complete %>%
mutate(gyield_14 = GYdry/.94,
N_total = Plant_N_SI + Side_N_SI)Day 21 - 10/07/2024
Announcements
Model selection
- Recall the conceptual pipeline we use to solve our problems using statistics.
- Idea/Question/Objective
- Gathering data
- Building a statistical model (i.e., assumptions about the data generating process)
- Fitting the model to data
- Model diagnostics
- Model selection
- Inference
- Communicating results
- Idea/Question/Objective
- Sometimes, there exists a group of models that could possibly answer our question in (1) and make relatively reasonable assumptions (i.e., they pass the test in (2)). So, we have to select between this group of models which one is the best.
- How many variables should we include?
- Bias-variance trade off
- We have to define what “good” means!
- Penalized fit (i.e., penalty for including too many parameters)
- Information Criteria (e.g., AIC, BIC)
- R2adj
- Information Criteria (e.g., AIC, BIC)
- Out-of-sample prediction accuracy
- Cross-validation
- RMSEP (prediction root mean squared error)
- Cross-validation
- A paper on how to evaluate predictions versus observed values.
- Penalized fit (i.e., penalty for including too many parameters)
Applications
Using the data from Ransom et al. (2021), we wish to create a function that describes the relationship between Nitrogen supply and corn yield. Firstly, we will focus on a single site and year (Ames, IA, 2014).
Load the data
dd_complete %>%
ggplot(aes(N_total, gyield_14))+
geom_point()+
theme_classic()+
labs(x = expression(Total~N~Applied~(kg~ha^{-1})),
y = expression(Grain~Yield~(kg~ha^{-1})))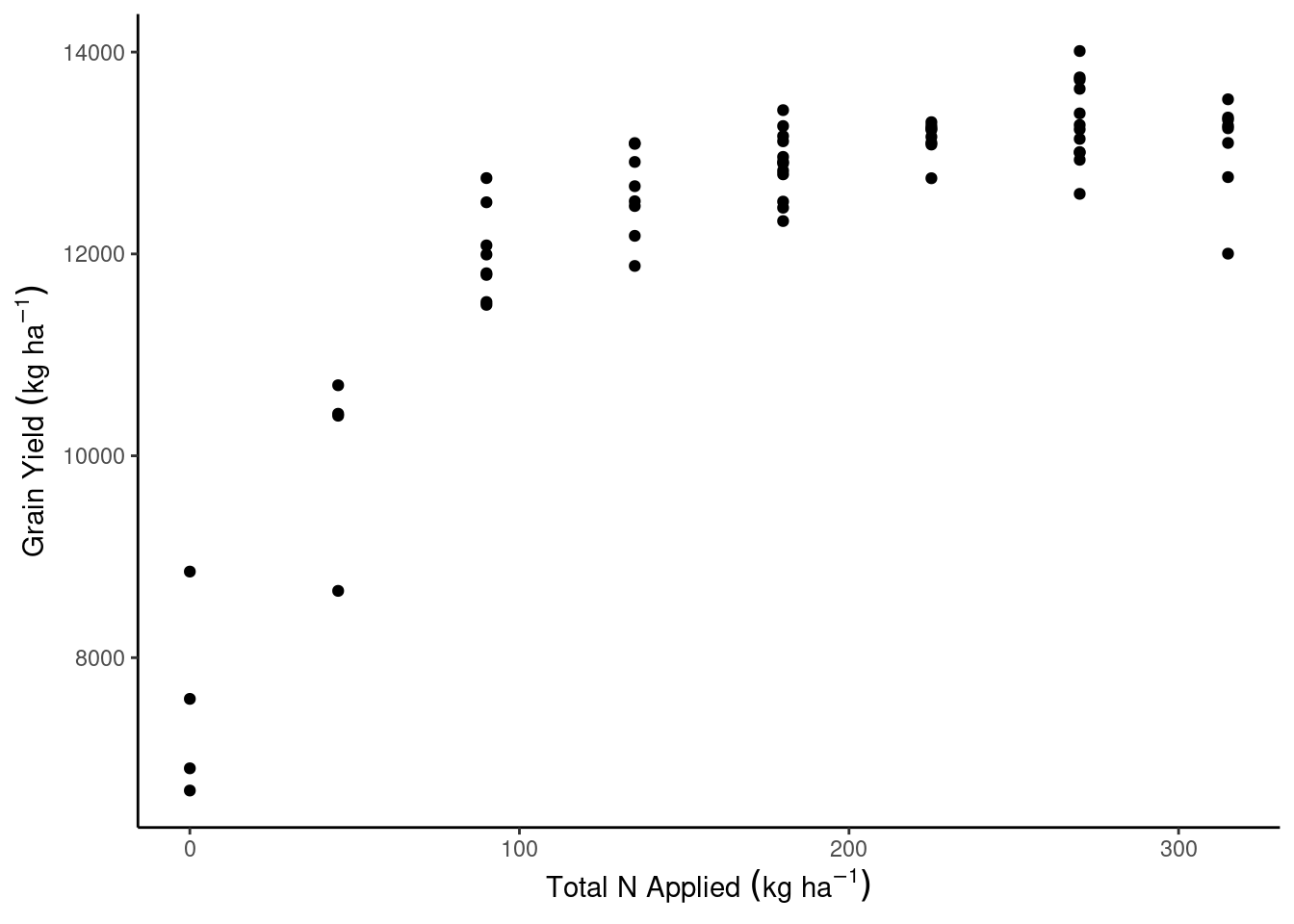
Fit the to some models
m0 <- lm(gyield_14 ~ N_total, data = dd_complete)
m1 <- lm(gyield_14 ~ N_total + I(N_total^2), data = dd_complete)
m2 <- lm(gyield_14 ~ N_total + I(N_total^2) + Block , data = dd_complete)
m3 <- lm(gyield_14 ~ N_total + I(N_total^2) + Block + PMPM, data = dd_complete)
m4 <- lm(gyield_14 ~ N_total + I(N_total^2) + Block + PMPM + Clay, data = dd_complete)
m5 <- lm(gyield_14 ~ N_total + I(N_total^2) + Block + PMPM + Clay + Sand, data = dd_complete)
m6 <- lm(gyield_14 ~ N_total + I(N_total^2) + Block + PMPM + Clay + Sand + Silt, data = dd_complete)n <- nobs(m1)Select the best models
AIC
\[AIC_M = 2p - 2\log(\hat{L}),\]
p <- length(coef(m1))+1
log_lik <- -n / 2 * (log(2 * pi) + log(sum(m1$residuals^2) / n) + 1)
2*p - 2*log_lik[1] 997.5004AIC(m1)[1] 997.5004BIC
\[BIC_M = p \cdot log(n) - 2\log(\hat{L}),\]
# for BIC, k is log(nobs(m1))
log(nobs(m1))*p - 2*log_lik[1] 1006.136AIC(m1, k = log(nobs(m1)))[1] 1006.136BIC(m1)[1] 1006.136R2 and R2adj
Recall that
\[R^2 = 1-\frac{SSE}{SST}\] is considered the proportion of variation in y explained by variation in x (or in my model, if I have more than one predictor).
R2, however, does not penalize for adding more predictors (i.e., more parameters) and can only increase. We solve this by calculating
\[R^2_{adj} = R^2 - (1 - R^2) \frac{p-1}{n-p},\] where \(n\) is the number of observations and \(p\) is the number of parameters.
Downfalls of using R2 for model selection
- Uncertainty (precision is tricky with small sample size)
- Using the same data twice (once to fit the models, then to judge the models!)
- Anscombe’s quartet
A summary
summary(m5)
Call:
lm(formula = gyield_14 ~ N_total + I(N_total^2) + Block + PMPM +
Clay + Sand, data = dd_complete)
Residuals:
Min 1Q Median 3Q Max
-1358.67 -369.15 50.32 395.71 1328.61
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.785e+04 2.108e+04 -0.847 0.401
N_total 4.556e+01 2.838e+00 16.052 <2e-16 ***
I(N_total^2) -9.704e-02 8.118e-03 -11.955 <2e-16 ***
Block 4.780e+02 3.068e+02 1.558 0.125
PMPM 1.999e+01 1.962e+01 1.019 0.313
Clay 6.088e+02 5.213e+02 1.168 0.248
Sand 2.290e+02 2.007e+02 1.141 0.259
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 554.9 on 57 degrees of freedom
Multiple R-squared: 0.8856, Adjusted R-squared: 0.8736
F-statistic: 73.55 on 6 and 57 DF, p-value: < 2.2e-16summary(m6)
Call:
lm(formula = gyield_14 ~ N_total + I(N_total^2) + Block + PMPM +
Clay + Sand + Silt, data = dd_complete)
Residuals:
Min 1Q Median 3Q Max
-1358.67 -369.15 50.32 395.71 1328.61
Coefficients: (1 not defined because of singularities)
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.785e+04 2.108e+04 -0.847 0.401
N_total 4.556e+01 2.838e+00 16.052 <2e-16 ***
I(N_total^2) -9.704e-02 8.118e-03 -11.955 <2e-16 ***
Block 4.780e+02 3.068e+02 1.558 0.125
PMPM 1.999e+01 1.962e+01 1.019 0.313
Clay 6.088e+02 5.213e+02 1.168 0.248
Sand 2.290e+02 2.007e+02 1.141 0.259
Silt NA NA NA NA
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 554.9 on 57 degrees of freedom
Multiple R-squared: 0.8856, Adjusted R-squared: 0.8736
F-statistic: 73.55 on 6 and 57 DF, p-value: < 2.2e-16metrics <- data.frame(model = c("N_total",
"N_total + I(N_total^2)",
"N_total + I(N_total^2) + Block",
"N_total + I(N_total^2) + Block + PMPM",
"N_total + I(N_total^2) + Block + PMPM + Clay",
"N_total + I(N_total^2) + Block + PMPM + Clay + Sand",
"N_total + I(N_total^2) + Block + PMPM + Clay + Sand + Silt"),
R2 = c(summary(m0)$r.squared, summary(m1)$r.squared, summary(m2)$r.squared,
summary(m3)$r.squared, summary(m4)$r.squared, summary(m5)$r.squared, summary(m6)$r.squared),
R2_adj = c(summary(m0)$adj.r.squared, summary(m1)$adj.r.squared, summary(m2)$adj.r.squared,
summary(m3)$adj.r.squared, summary(m4)$adj.r.squared, summary(m5)$adj.r.squared,
summary(m6)$adj.r.squared),
AIC = AIC(m0, m1, m2, m3, m4, m5, m6)$AIC,
BIC = BIC(m0, m1, m2, m3, m4, m5, m6)$BIC) %>%
mutate(across(R2:BIC, ~round(., 3)))
knitr::kable(metrics, format = "html")| model | R2 | R2_adj | AIC | BIC |
|---|---|---|---|---|
| N_total | 0.589 | 0.582 | 1070.935 | 1077.412 |
| N_total + I(N_total^2) | 0.873 | 0.869 | 997.500 | 1006.136 |
| N_total + I(N_total^2) + Block | 0.878 | 0.872 | 997.147 | 1007.941 |
| N_total + I(N_total^2) + Block + PMPM | 0.882 | 0.874 | 996.822 | 1009.775 |
| N_total + I(N_total^2) + Block + PMPM + Clay | 0.883 | 0.873 | 998.453 | 1013.566 |
| N_total + I(N_total^2) + Block + PMPM + Clay + Sand | 0.886 | 0.874 | 999.008 | 1016.279 |
| N_total + I(N_total^2) + Block + PMPM + Clay + Sand + Silt | 0.886 | 0.874 | 999.008 | 1016.279 |
data_plot <- expand.grid(N_total = seq(0, 300, by = 5),
Block = 1:4)
data_plot <- data_plot %>% bind_cols(predict(m2, newdata = data_plot, interval = "confidence"))dd_complete %>%
ggplot(aes(N_total, gyield_14))+
theme_classic()+
geom_ribbon(aes(y = fit, ymin = lwr, ymax = upr, group = Block), data = data_plot, alpha = .15)+
geom_line(aes(y = fit, group = Block), data = data_plot)+
geom_point()+
labs(x = expression(Total~N~Applied~(kg~ha^{-1})),
y = expression(Grain~Yield~(kg~ha^{-1})))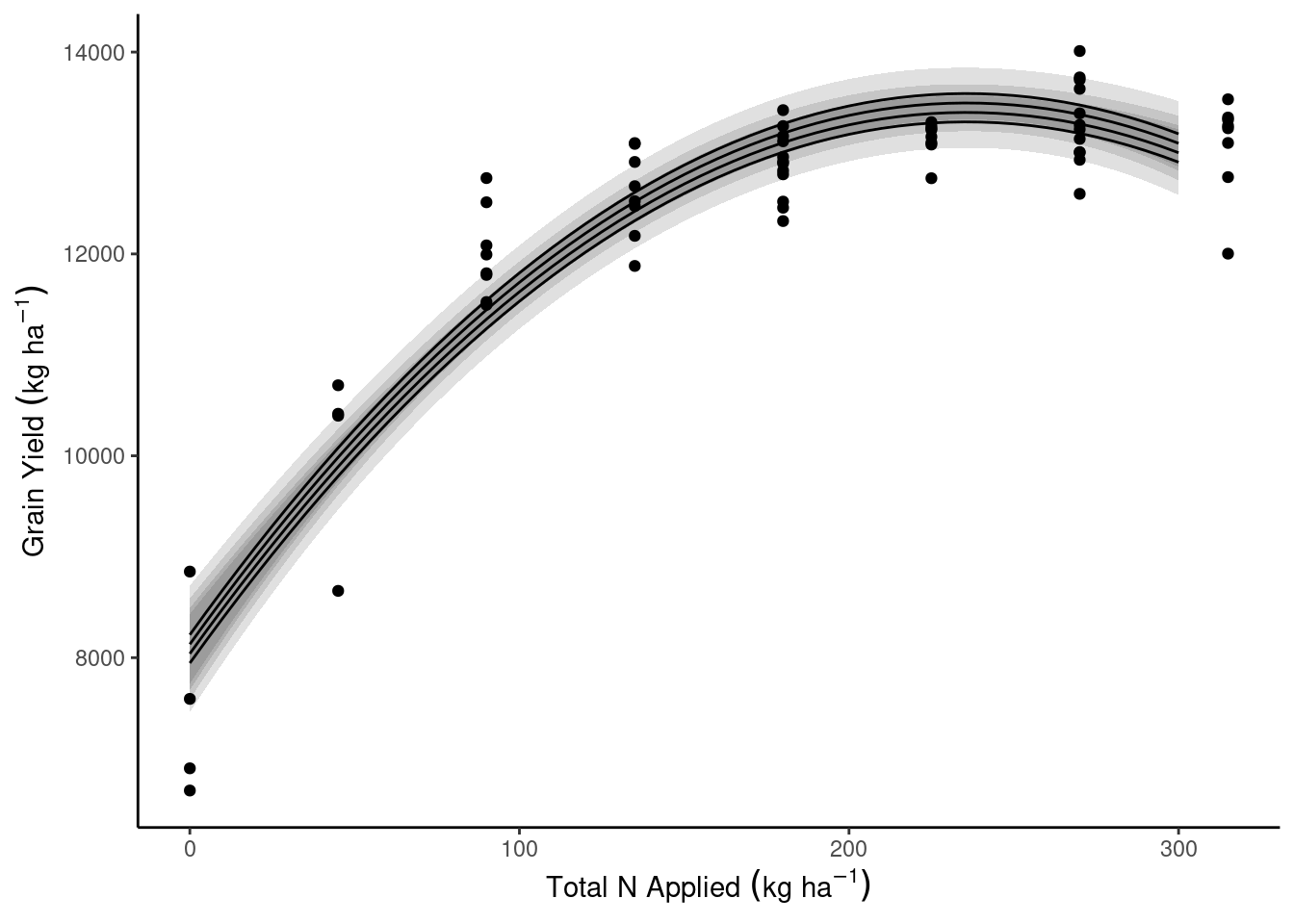
A question for the class
How do you expect the variable selection to change if we add more…
- sites
- years
Important questions/concepts
Variable selection - checkpoints
- How do the predictions compare?
- Is the data generation process well represented?
- What is the cost/benefit relationship of including certain predictors?
Watch out with
- Collinearity
- Using the same data twice (once to fit the models, then to judge the models!)
For next class
- Read Chapter 8 & 10
- Finish Assignment 3